Mapping the energy landscape for second stage folding of a single membrane protein
A collaboration between Bowie Laboratory and a research team led by Tae-Young Yoon of the Department of Physics at the Korea Advanced Institute of Science and Technology has employed the use of a magnetic tweezer to “quantitatively map the folding energy landscape, the folding kinetic rate, and folding intermediates of a membrane protein in a membrane environment for the first time.” (Phys.org)
Members of Bowie lab have provided the following abstract and external links:
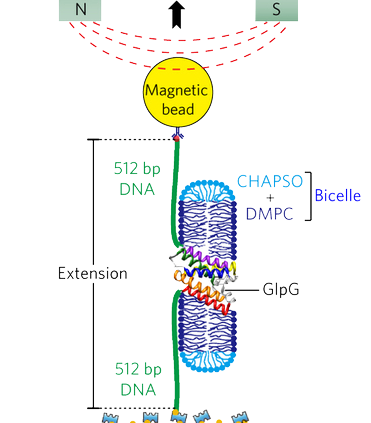
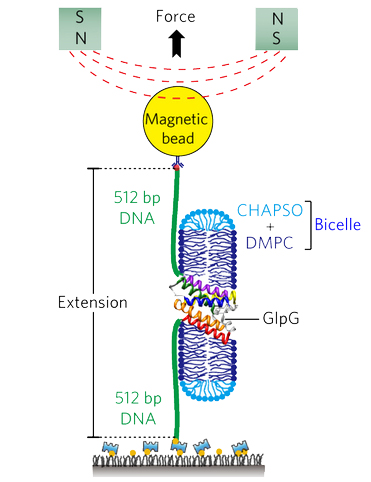
Membrane proteins are designed to fold and function in a lipid membrane, yet folding experiments within a native membrane environment are challenging to design. Here we show that single molecule forced unfolding experiments can be adapted to study helical membrane protein folding under native-like bicelle conditions. Applying force using magnetic tweezers, we find that a transmembrane helix protein, E. coli rhomboid protease GlpG, unfolds in a highly cooperative manner, largely unraveling as one physical unit in response to mechanical tension above 25 pN. Considerable hysteresis is observed, with refolding occurring only at forces below 5 pN. Characterizing the energy landscape reveals only modest thermodynamic stability (DG = 6.5 kBT) but a large unfolding barrier (21.3 kBT) that can maintain the protein in a folded state for long periods of time (t1/2 ~3.5 hrs). The observed energy landscape may have evolved to limit the existence of troublesome partially unfolded states and impart rigidity to the structure.
Phys.org
http://phys.org/news/2015-10-membrane-protein.html
Nature Chemical Biology
http://www.nature.com/nchembio/journal/vaop/ncurrent/full/nchembio.1939.html