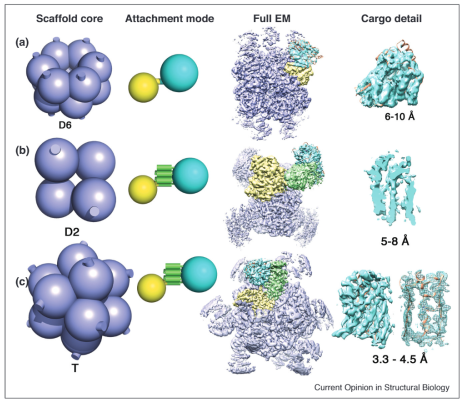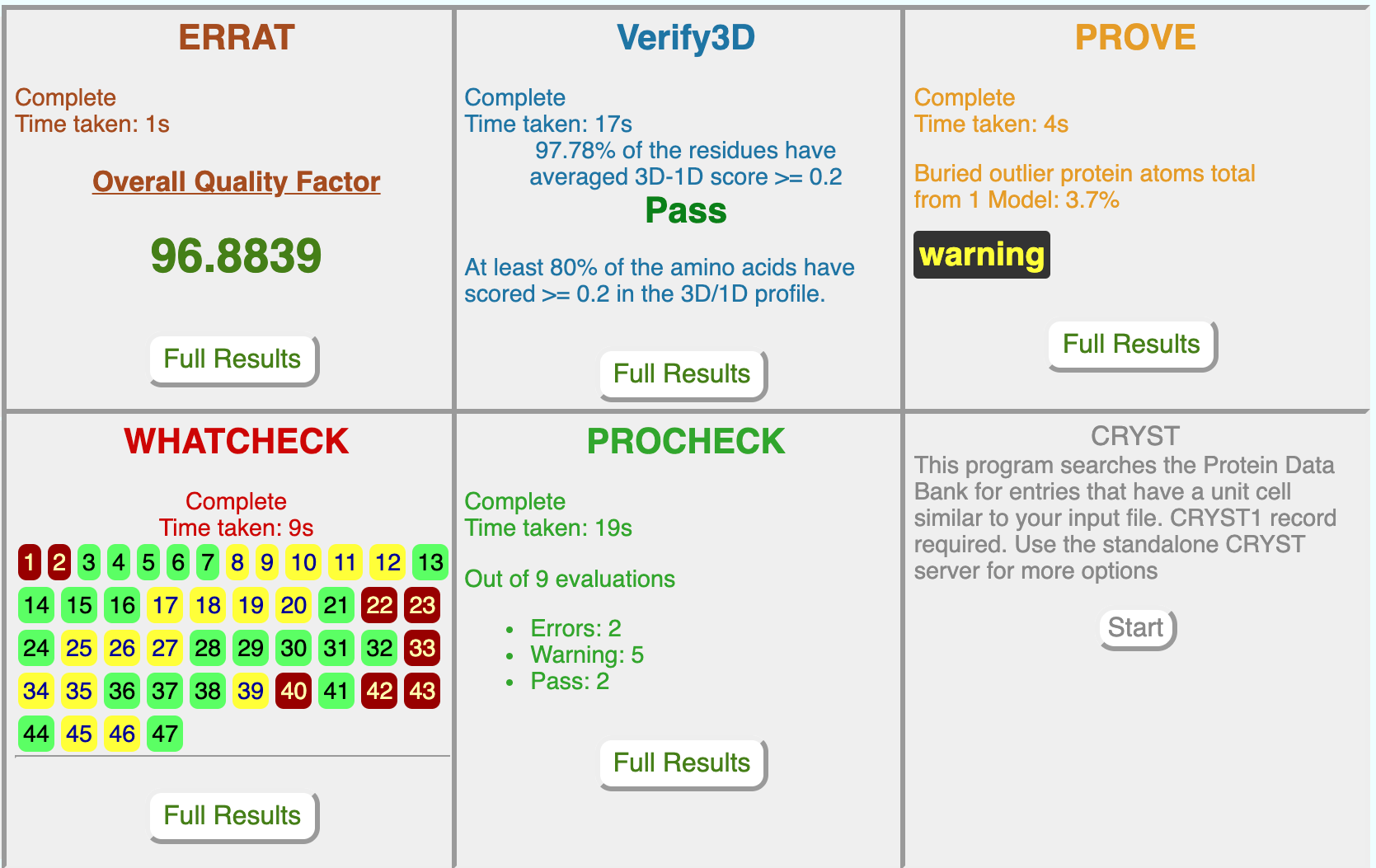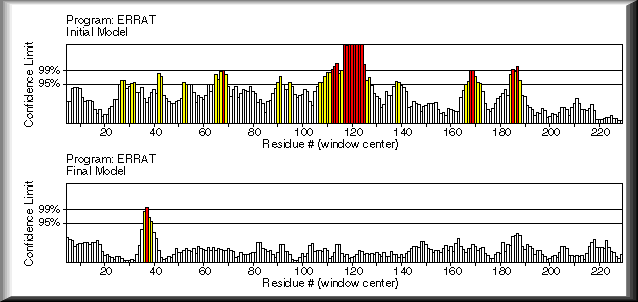
ERRAT is a program for verifying protein structures determined by crystallography. Error values are plotted as a function of the position of a sliding 9-residue window. The error function is based on the statistics of non-bonded atom-atom interactions in the reported structure (compared to a database of reliable high-resolution structures).
The figure shows a plot of an initial model and a final model. Regions of the structure that can be rejected at the 95% confidence level are yellow; 5% of a good protein structure is expected to have an error value above this level. Regions that can be rejected at the 99% level are shown in red. According to the analysis by ERRAT, the final model is significantly improved relative to the initial model.
Generally speaking, the method is sensitive to smaller errors than 3-D Profile analysis (Bowie & Eisenberg), but is more forgiving than Procheck (Thornton). For a recent review of protein verification algorithms, see MacArthur, et al. Curr. Opin. Struct. Biol. 4, 731-737 (1994).
- Abstract of the ERRAT paper by Colovos and Yeates.
- Retrieve the C++ code as a tar file.
- Go to the SAVES server to run ERRAT.