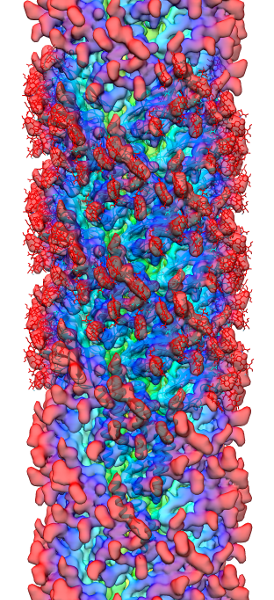
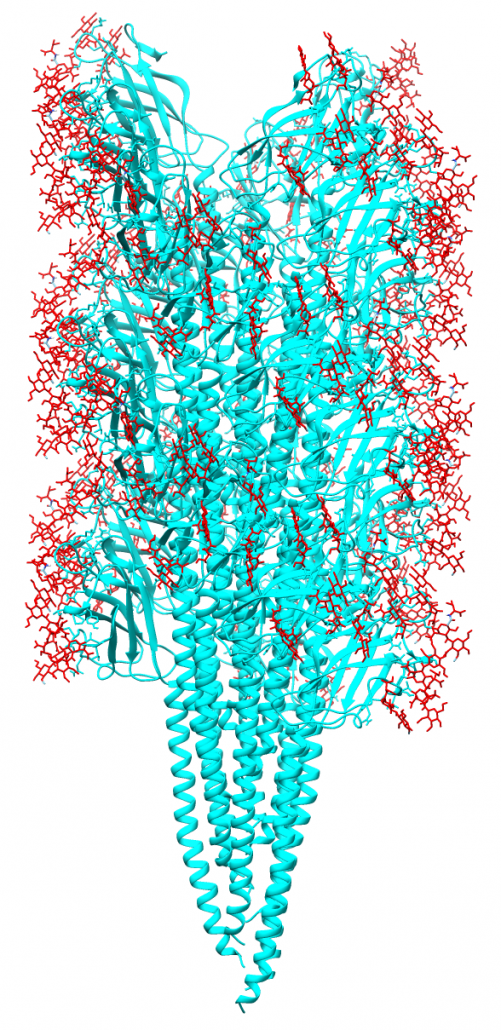
Methanospirillum hungatei cells and flagella structure
The archaeal motility structure (flagellum /archaellum)
During an initial structural and proteomic survey of the model archaeal microbe Methanospirillum hungatei we identified proteins comprising the attached cellular flagellum. Little was previously known about these archaeal appendages and we therefore characterized them and obtained the first atomic-level structure of a model archaeal flagellum. The findings reveal properties distinct from the bacterial flagellar counterpart regarding subunit size, post translational modifications and tertiary structure. Experiments are continuing to further characterize the post translational glycan modifications to the M. hungatei flagellum in collaborations by the Gunsalus and Loo Laboratories of the UCLA-DOE Institute.
Genome-wide transcriptomics in Chlamydomonas reinhardtii during day-night cycles reveals new metabolic patterns in light and stress response (Merchant lab)
A day in the life of Chlamydomonas
The unicellular green alga Chlamydomonas reinhardtii excels at acclimating to a changing environment. We analyzed expression patterns of its three genomes in cells grown under light-dark cycles. Nearly 85% of transcribed genes show differential expression, with different sets of transcripts being up-regulated over the course of the day to coordinate cellular growth before undergoing cell division. Parallel measurements of select metabolites and pigments, physiological parameters and a subset of proteins allow us to infer metabolic events and to evaluate the impact of the transcriptome on the proteome. Among new findings is the observation that Chlamydomonas exhibits low respiratory activity at night and relies instead on fermentative metabolism; we propose that the ferredoxin FDX9 acts as the electron donor to fermentative hydrogenases. The light stress responsive genes PSBS, LHCSR1 and LHCSR3 all show an acute response to light at dawn under abrupt dark-to light transitions. LHCSR3 genes also exhibits a later burst in expression in the middle of the day in response to higher light intensities. Each response to light (acute and sustained) can be selectively activated under specific conditions. Our expression dataset, complemented with co-expression networks and metabolite profiling, should constitute an excellent resource for the algal and plant communities.
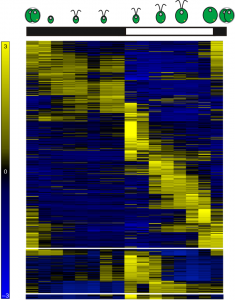
Figure 1: Heatmap representing gene expression for 11,306 differentially expressed genes over the diurnal cycle. Chlamydomonas cells grown under light-dark cycles accumulate biomass as a direct consequence of photosynthesis during the day, and divide shortly after dusk.
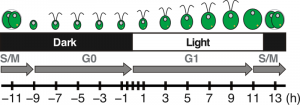
Figure 2: Diagram of Chlamydomonas cell division progression over the diurnal cycle.
Cells grow in volume and accumulate biomass during the day, using photosynthesis to fuel growth. Around dusk, cells will initiate the mitotic division and yield two daughter cells per mother cell. At night, daughter cells will regenerate their flagella, which were resorbed before cell division, and enter the quiescent phase of the cell cycle until the following dawn.
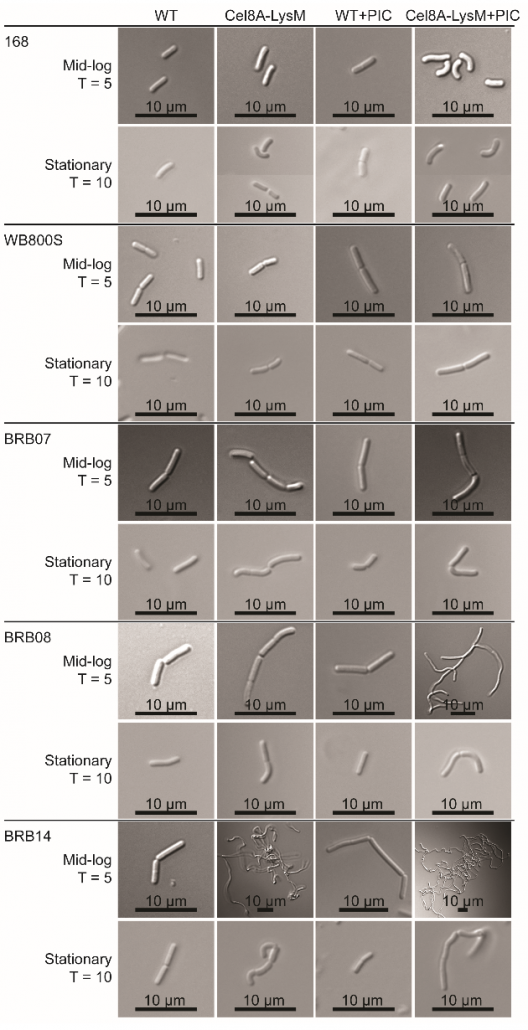
New method to create stable enzyme coated B. subtilis cells for biotechnological applications.
Microbes engineered to display heterologous proteins could be useful biotechnological tools for protein engineering, lignocellulose degradation, biocatalysis, bioremediation and biosensing. Bacillus subtilis is a promising host to display proteins, as this model Gram-positive bacterium is genetically tractable and already used industrially to produce enzymes. The Clubb group developed a unique two-step procedure that enables the construction of enzyme coated vegetative B. subtilis cells that retain stable cell-associated enzyme activity for nearly 3 days. The results of this work could aid the development of whole cell display systems that have useful biotechnological applications.
Huang GL, Gosschalk JE, Kim YS and Clubb RT. Stabilizing displayed proteins on vegetative Bacillus subtilis cells. Applied Microbiology and Biotechnology 102 2018; 6547-6565